Timing in R
Published: November 20, 2017

As time goes on, your R scripts are probably getting longer and more complicated, right? Timing parts of your script could save you precious time when re-running code over and over again. Today I’m going to go through the 4 main functions for doing so.
Nested timings
1) Sys.time()
Sys.time() takes a “snap-shot” of the current time and so it can be used to record start and end times of code.
start_time = Sys.time()
Sys.sleep(0.5)
end_time = Sys.time()
To calculate the difference, we just use a simple subtraction
end_time - start_time
## Time difference of 0.5027 secs
Notice it creates a neat little message for the time difference.
2) The {tictoc} package
You can install the CRAN version of {tictoc} via
install.packages("tictoc")
whilst the most recent development is available via the {tictoc} GitHub page.
library("tictoc")
Like Sys.time(), {tictoc} also gives us ability to nest timings within code. However, we now have some more options to customise our timing. At it’s most basic it acts like Sys.time():
tic()
Sys.sleep(0.5)
toc()
## 0.505 sec elapsed
Now for a more contrived example.
# start timer for the entire section, notice we can name sections of code
tic("total time")
# start timer for first subsection
tic("Start time til half way")
Sys.sleep(2)
# end timer for the first subsection, log = TRUE tells toc to give us a message
toc(log = TRUE)
## Start time til half way: 2.013 sec elapsed
Now to start the timer for the second subsection
tic("Half way til end")
Sys.sleep(2)
# end timer for second subsection
toc(log = TRUE)
## Half way til end: 2.005 sec elapsed
# end timer for entire section
toc(log = TRUE)
## total time: 4.027 sec elapsed
We can view the results as a list (format = TRUE returns this list in a nice format), rather than raw code
tic.log(format = TRUE)
## user system elapsed
## 0.000 0.000 1.001
We only want to take notice of the “elapsed” time, for the definition of the “user” and “system” times see this thread.
For a repeated timing, we would use the replicate() function.
system.time(replicate(10, Sys.sleep(0.1)))
## user system elapsed
## 0.004 0.000 1.004
2) The microbenchmark package
You can install the CRAN version of {microbenchmark} via
install.packages("microbenchmark")
Alternatively you can install the latest update via the {microbenchmark} GitHub page.
library("microbenchmark")
At it’s most basic, microbenchmark() can we used to time single pieces of code.
# times = 10: repeat the test 10 times
# unit = "s": output in seconds
microbenchmark(Sys.sleep(0.1), times = 10, unit = "s")
## Unit: seconds
## expr min lq mean median uq max neval
## Sys.sleep(0.1) 0.1001 0.1002 0.1002 0.1002 0.1002 0.1002 10
Notice we get a nicely formatted table of summary statistics. We can record our times in anything from seconds to nanoseconds(!!!!). Already this is better than system.time(). Not only that, but we can compare sections of code in an easy-to-do way and name the sections of code for an easy-to-read output.
sleep = microbenchmark(sleepy = Sys.sleep(0.1),
sleepier = Sys.sleep(0.2),
sleepiest = Sys.sleep(0.3),
times = 10,
unit = "s")
As well as this (more?!) {microbenchmark} comes with a two built-in plotting functions.
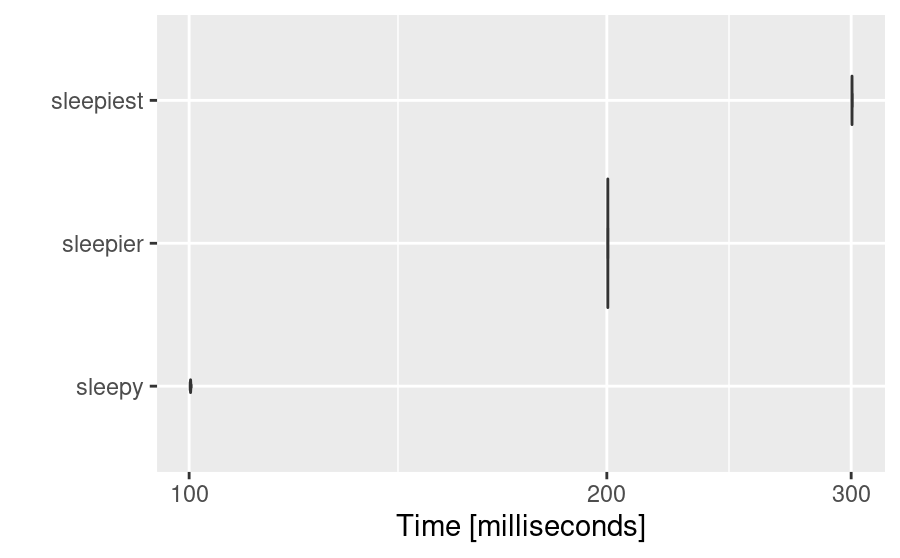
microbenchmark:::autoplot.microbenchmark(sleep)
microbenchmark:::boxplot.microbenchmark(sleep)
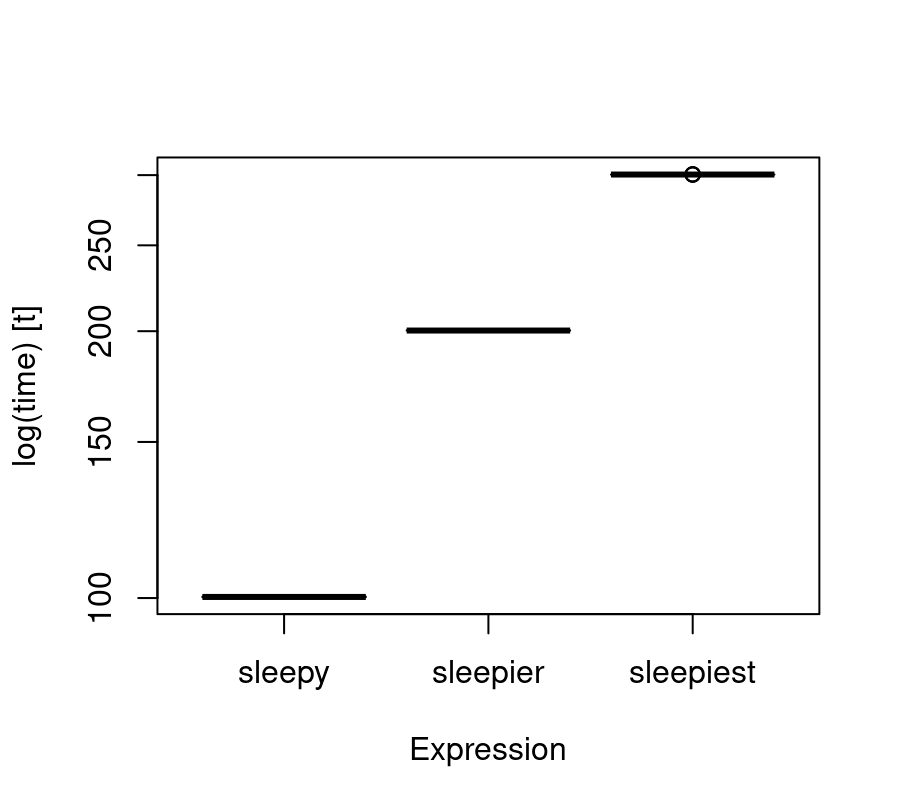
These provide quick and efficient ways of visualising our timings.
Conclusion
Sys.time() and system.time() have there place, but for most cases we can do better. The {tictoc} and {microbenchmark} packages are particularly useful and make it easy to store timings for later use, and the range of options for both packages stretch far past the options for Sys.time() and system.time(). The built-in plotting functions are handy.
Thanks for chatting!

